Nil Aygün led a study using causal inference tools to infer how genetic variants influence gene expression through changes in chromatin accessibility and through changes in expression of other genes. This allows an understanding of how genetic variation leads to changes in gene expression within specific cell types present during neurogenesis. This study is available as a pre-print here.
Using genetics to understand how evolution shaped modern human brain structure
Led by the lab of Simon Fisher, we attempted to replicate some of our previous findings on how evolution has changed the size and shape of the human brain. We found that our previous evidence of recent selection influencing cortical structure was not replicated in an independent sample. But, human gained enhancers did have a replicable effect on human brain structure. This study was published in PNAS.
MPRA review published
Jessica McAfee led a review describing the many different types of multiplex reporter assays and how they will be useful for finding causal genetic variants associated with brain traits. This work was in collaboration with the lab of Hyejung Won. The study was published in the Journal of Developmental Disorders.
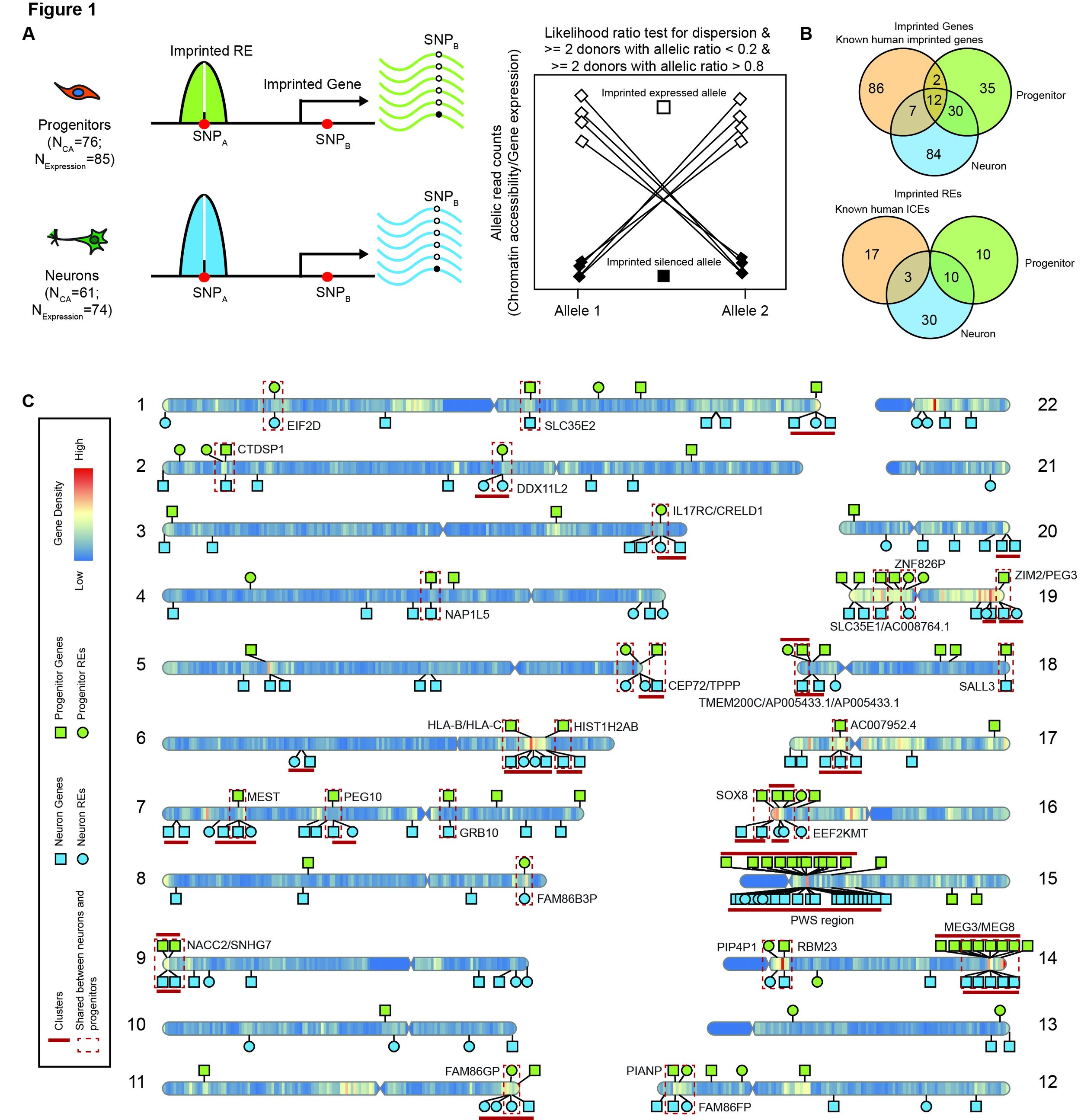
Inferring imprinting during human neurogenesis
Dan Liang led a study inferring cell-type specific imprinting during human neurogenesis. This study was published at Human Molecular Genetics. These sites may be particularly useful to interpret parent of origin genetic association studies, as they start getting well powered.
JOVE paper on tissue clearing and analysis methods
Felix Kyere and Ian Curtin led a paper describing how our lab performs MRI, tissue clearing, and analysis in order to understand how genetic variations influence brain structure at gross volumetric and cellular levels. Contributions to this paper are from several lab members including Carolyn McCormick, and collaborations with the lab of Guorong Wu and Ian Shih. The paper is available here.
Book chapter published on organoids
Rose Glass and Felix Kyere published a book chapter within the book entitled “Phenotyping of Human iPSC-derived Neurons”. The chapter is entitled “Brain organoids: models of cell type diversity, connectivity, and disease phenotypes.” The chapter covers some of the advantages and disadvantages of organoids as a model system, along with some recommendations for reproducible phenotypes and characterization. This chapter was written together with colleagues at CHOP - Elisa Waxman and Deb French.
Multiple graduate students win funding
Jordan Valone (Bioinformatics and Computational Biology), Ian Curtin (Genetics and Molecular Biology and Certificate Program in Translational Medicine), and former Stein lab technician and current Won lab graduate student Jessica McAfee (Genetics and Molecular Biology) all won NIH T32 training grants to support their graduate research. Congrats all!
Nil Aygün wins Terry Magnuson Award
Bioinformatics and Computational Biology graduate student Nil Aygün won the Terry Magnuson award for Outstanding Graduate Student at the 2022 Genetics Retreat. Congrats, Nil!
Stein lab at World Congress of Psychiatric Genetics 2022
Nana Matoba and Jordan Valone attended the World Congress of Psychiatric Genetics 2022 in Florence, Italy. Nana Matoba won the WCPG Early Career Investigator award.
Stein lab at International Society for Autism Research 2022
Justin Wolter, Rose Glass, and Felix Kyere attended INSAR 2022 in Austin, TX. Rose and Felix won travel awards to attend the conference. Rose and Justin both gave talks on their work and Felix presented a poster.
Deep discussions on brain development and/or typos.
Mike Lafferty successfully defends his dissertation
Dr. Mike Lafferty successfully defended his dissertation in March 2022, where he discovered genetic variants modulating levels of miRNAs in the developing human cortex. Mike was in the Bioinformatics and Computational Biology curriculum and is the 3rd PhD from our lab.
Mike receives the Paul Hollywood handshake for excellent dissertation.
Mike was kind enough to share his pizza with the group.
It was a very sunny day!
Molecular Psychiatry 2022 Conference
Many members of the lab attended and presented at the Molecular Psychiatry 2022 conference. Postdoc Nana Matoba won a Young Investigator Award. Jason co-led a symposium on “Cell-type and context-specific effects of genetic risk variants for neuropsychiatric disorders” together with Alexi Nott, Hyejung Won, and Xin Jin (pictured below). Rose Glass gave a talk on modeling brain overgrowth in autism using cortical organoids.
Stein lab at American Society of Human Genetics 2021
Neuroscience graduate student Felix Kyere was a predoctoral finalist of the Charles J. Epstein Trainee Award for Excellence in Human Genetics Research. He gave a talk at the ASHG 2021 virtual conference on tissue clearing in a mouse model of autism. See an image from his talk in the tweet below. Nana Matoba also won the Revewers’ Choice award for her poster at this conference.
Neuroscientists Talk Shop Podcast
After giving a talk at University of Texas San Antonio, Jason talks with fellow neuroscientists Melanie Carless and Salma Quraishi about genetics, causality, brain organoid models, and large scale studies of the genetics of brain structure and disease.
Jason promoted to Associate Professor
Effective January 1, 2022, Jason is promoted to associate professor with tenure. I feel very humbled and grateful to all of the hardworking people in my lab, and all of my previous and current mentors. What a ride!
Nuclei Ninja party
The nuclei ninjas all gathered in person for some pizza, a lab tour, and some career advice. The nuclei ninjas (see nucleininja.org) are undergraduates who are creating a manually labeled training dataset to train our nuclear recognition algorithms. Thanks for all your hard work you guys!
NuMorph published in Cell Reports
We developed NuMorph, a pipeline to analyze tissue cleared mouse brain images. NuMorph was applied to two different mouse models, finding the cell types underlying gross structural brain differences. From beautiful images to quantifications to biological insights! This work was led by biomedical engineering graduate student, Oleh Krupa. Check out the manuscript here.
Preprint on imprinting during neuronal differentiation
In our new preprint, we identify putatively imprinted regions and genes in cultured progenitors and differentiated neurons, finding many of them are cell-type specific. Our hope is that these genes and elements can be used together with parent of origin association studies to explain mechanisms of associated variants.
e/sQTL paper published in the american journal of human genetics
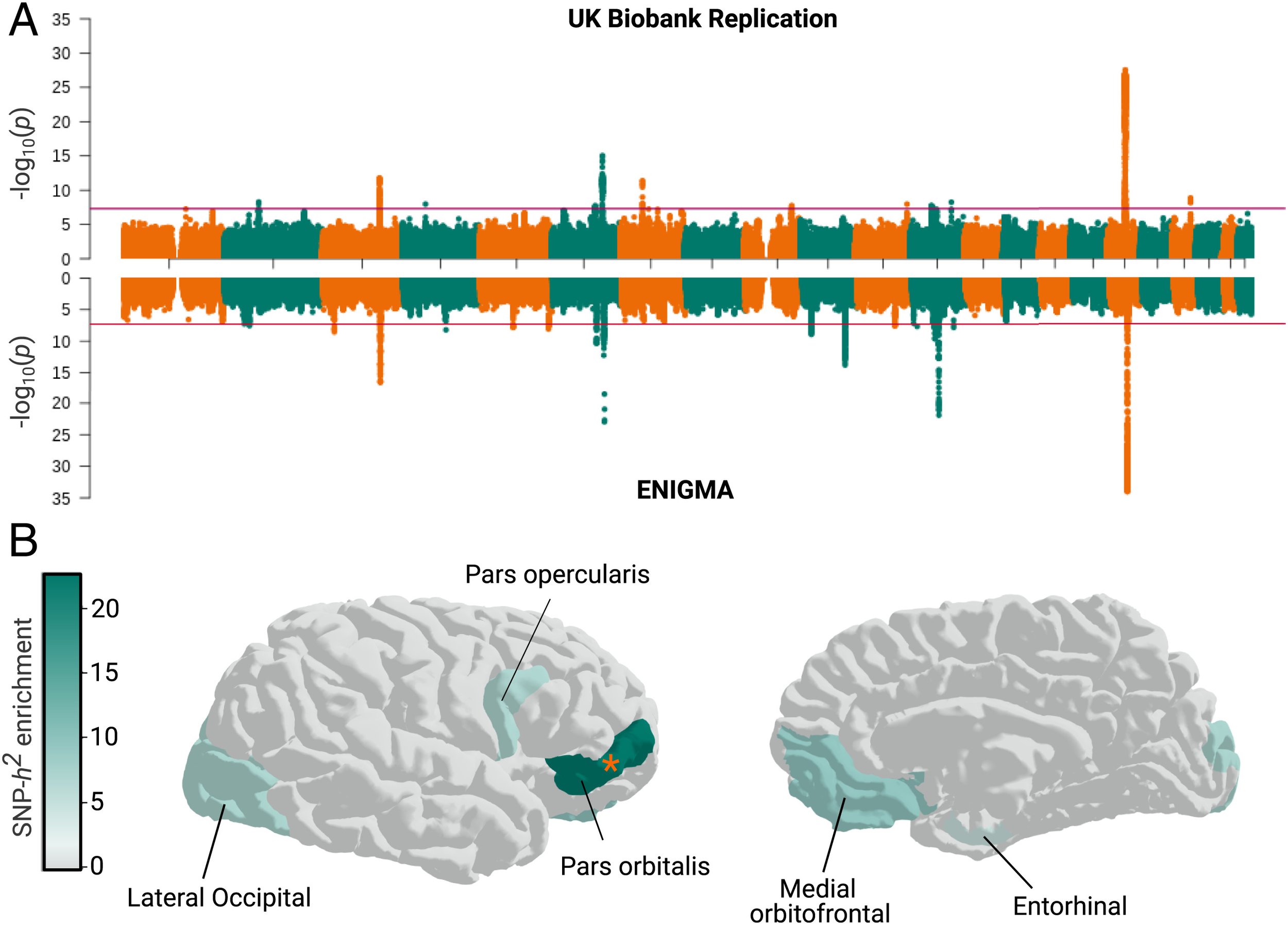
Your DNA sequence is the same in every cell in your body, and is slightly different between any two individuals. Genetic differences can lead to changes in the size and structure of our brains, and risk for diseases. Despite that the DNA is identical in every cell in the body (with few exceptions), genetic variants can exert their effects only within specific cell types. In our paper, led by graduate student Nil Aygun, we mapped the effects of genetic variants on gene expression in two cell types of the developing cortex, neurons and progenitors, modeled with neural stem cells. We used our results to explain which genes and cell types influence brain structure. In the figure above, we found that a genetic variant affecting CENPW expression only in progenitors influences global cortical surface area. The results from this study can lead to a better understanding of how genetic variation leads to changes in our brains and risk for neuropsychiatric disorders.
Genetics of white matter tracts published in Science
In work led by Hongtu Zhu and Bingxin Zhao, we explored how genetic variation influences brain region connectivity. In a sample size of over 43,000 individuals, many novel loci associated with white matter tracts were detected. Interestingly, some tracts showed a genetic correlation to schizophrenia, finally providing a solid genetic correlation to this disorder using a brain imaging based phenotype. Also, genetic variants within regulatory elements of glia, but not neurons, strongly influence white matter tracts. This provides a way of understanding which cell types are influencing macroscale brain structure traits. You can read more about this here.