We developed NuMorph, a pipeline to analyze tissue cleared mouse brain images. NuMorph was applied to two different mouse models, finding the cell types underlying gross structural brain differences. From beautiful images to quantifications to biological insights! This work was led by biomedical engineering graduate student, Oleh Krupa. Check out the manuscript here.
Preprint on imprinting during neuronal differentiation
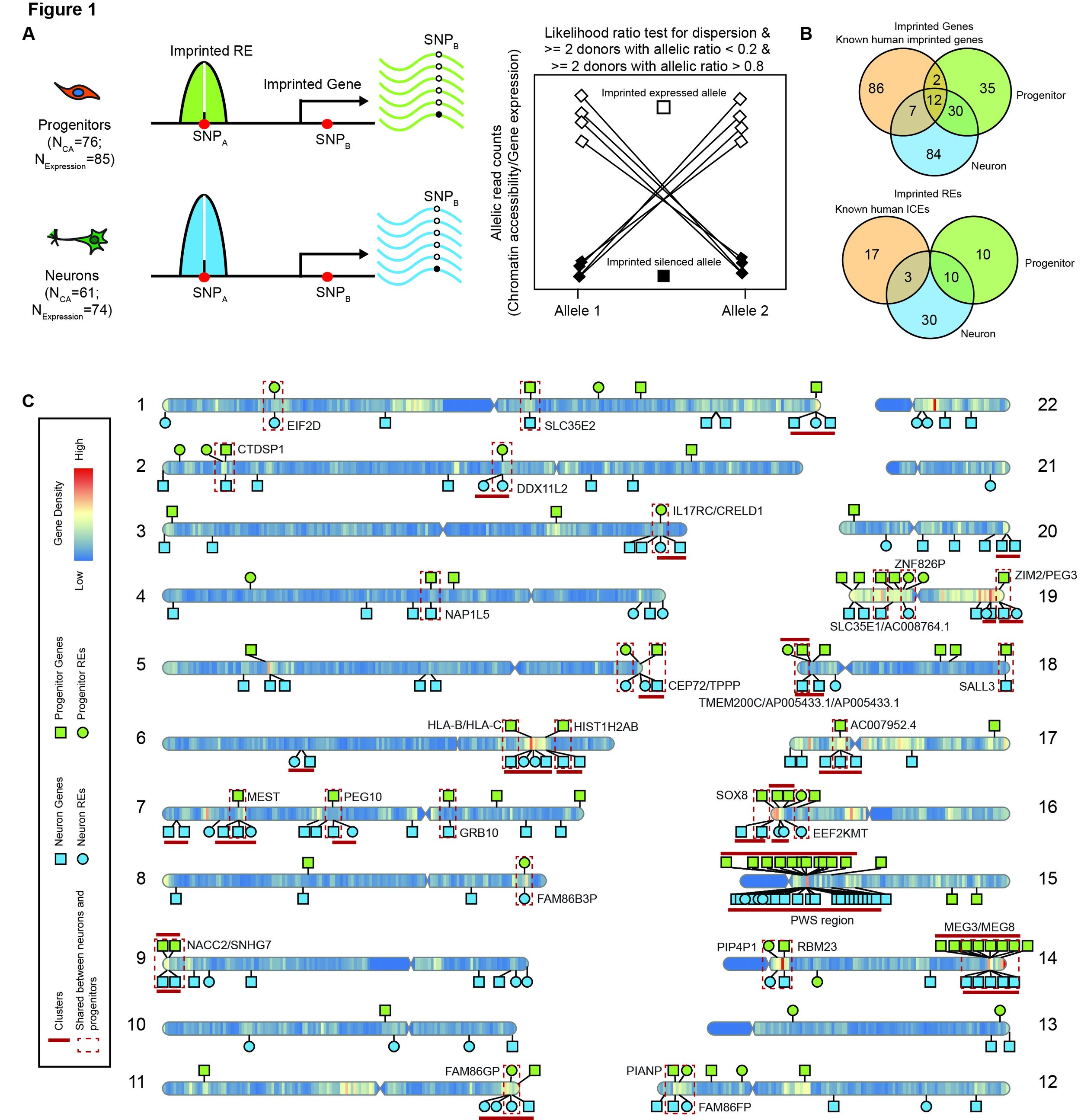
In our new preprint, we identify putatively imprinted regions and genes in cultured progenitors and differentiated neurons, finding many of them are cell-type specific. Our hope is that these genes and elements can be used together with parent of origin association studies to explain mechanisms of associated variants.
e/sQTL paper published in the american journal of human genetics
Your DNA sequence is the same in every cell in your body, and is slightly different between any two individuals. Genetic differences can lead to changes in the size and structure of our brains, and risk for diseases. Despite that the DNA is identical in every cell in the body (with few exceptions), genetic variants can exert their effects only within specific cell types. In our paper, led by graduate student Nil Aygun, we mapped the effects of genetic variants on gene expression in two cell types of the developing cortex, neurons and progenitors, modeled with neural stem cells. We used our results to explain which genes and cell types influence brain structure. In the figure above, we found that a genetic variant affecting CENPW expression only in progenitors influences global cortical surface area. The results from this study can lead to a better understanding of how genetic variation leads to changes in our brains and risk for neuropsychiatric disorders.
Genetics of white matter tracts published in Science
In work led by Hongtu Zhu and Bingxin Zhao, we explored how genetic variation influences brain region connectivity. In a sample size of over 43,000 individuals, many novel loci associated with white matter tracts were detected. Interestingly, some tracts showed a genetic correlation to schizophrenia, finally providing a solid genetic correlation to this disorder using a brain imaging based phenotype. Also, genetic variants within regulatory elements of glia, but not neurons, strongly influence white matter tracts. This provides a way of understanding which cell types are influencing macroscale brain structure traits. You can read more about this here.
Segment your 3D microscopy images with Segmentor
In order to train a computer what a nucleus looks like, you first have to provide manual annotations of features within microscopy images. This is difficult to do in 3D, so we developed a tool called Segmentor to make it easier and quicker. This will allow us to train our computers to recognize nuclei in 3D tissue cleared images. Read more about the tool here and download it at https://www.nucleininja.org/.
caQTL paper published in Nature Neuroscience
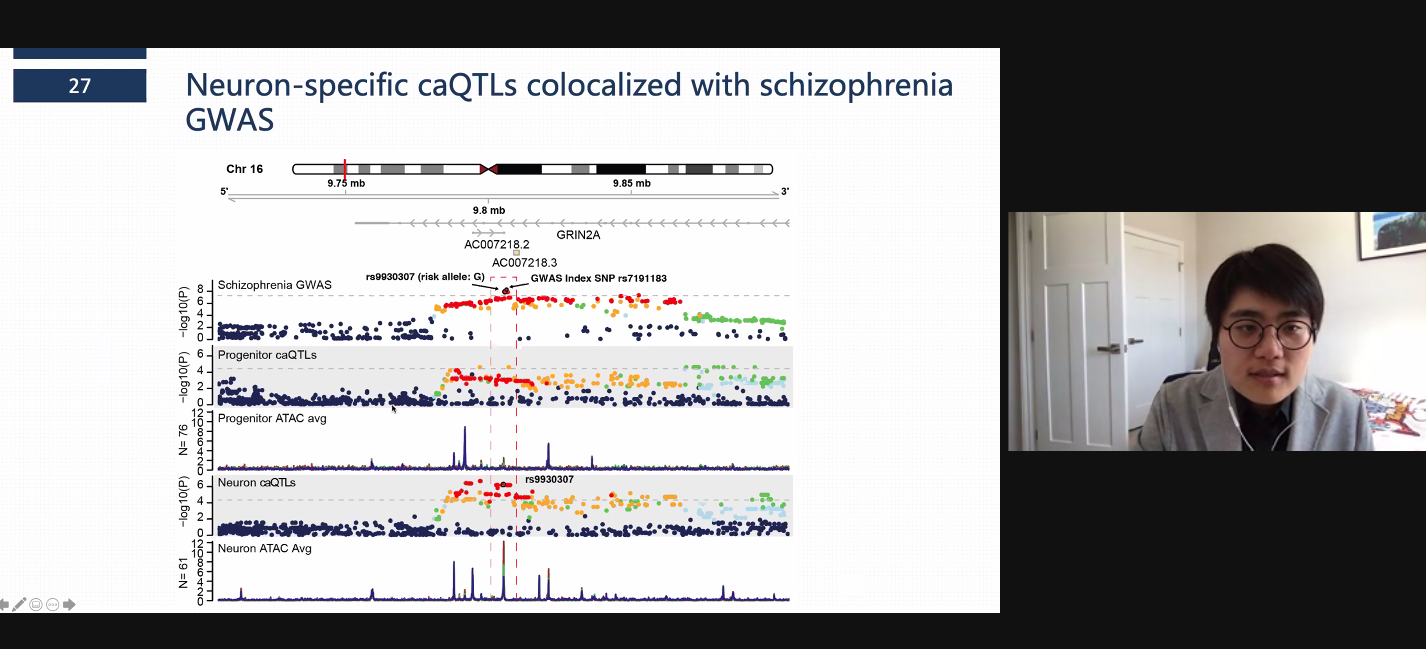
After many years of work, we published our cell-type specific chromatin accessibility paper in Nature Neuroscience. There were some interesting things we found: (1) caQTLs are highly cell-type specific, (2) caQTLs have higher effect sizes on average than eQTLs, (3) caQTLs can be used to identify causal variants, cell types, and mechanisms of non-coding GWAS loci. As an example, here is a locus associated with risk for schizophrenia near the GRIN2A gene where we show co-localization with a neuron-specific caQTL and allele specific chromatin accessibility, and experimentally validate the result with a luciferase assay. You can read the article here.
News & Views on UKBB Imaging Genomics in Nature Neuroscience
We wrote a News and Views in Nature Neuroscience on the expanded resource of Brain Imaging Genetics GWAS from the UK Biobank. We tried to frame what the advantages are to this new dataset and also suggest new directions for the field of imaging genetics, to better understand how genetic variation influences human brain structure and function. You can read more about it here.
Two new graduate students joining the lab
We welcome Jordan Valone (Bioinformatics and Computational Biology) and Ian Curtin (Genetics and Molecular Biology) to the lab. Jordan will work on stimulus specific genetic effects in brain development and Ian will work on brain structure changes in a mouse model of autism. Welcome aboard guys!
Dr. Oleh Krupa successfully defends his dissertation
Congratulations to Dr. Oleh Krupa, the second Ph.D. from the Stein lab! Oleh’s primary project was acquiring and analyzing tissue cleared images of brain structure (https://www.biorxiv.org/content/10.1101/2020.09.11.293399v2). He contributed to many other projects in the lab as well. We’re so proud of all you’ve accomplished!
Spotlight on Carla Escobar-Tomlienovich
Preprint released on Segmentor
We released a pre-print on Biorxiv about Segmentor, a tool for manual refinement of 3D nuclei annotations. Deep learning nuclear segmentation algorithms require a whole lot of manually labeled samples, both for training and for validation. So, we made a piece of software to make this process easier. The software is publicly available now at nucleininja.org.
Rose Glass awarded pre-doctoral NRSA
Congratulations to Neuroscience graduate student Rose Glass, who won a pre-doctoral NRSA! Her project will examine how well iPSC-derived cerebral organoids model the brain growth trajectory of the individuals from which they were made.
Justin Wolter leads study on potential gene therapy for Angelman syndrome
Congratulations to Justin Wolter (joint postdoc in the Stein and Zylka labs) and the entire Zylka lab for leading a study describing a CRISPR based gene therapy approach for increasing paternal UBE3A expression. This was validated not only in mouse, but also in neurons derived from primary human neural progenitors. The study was published in Nature.
Dr. Dan Liang successfully defends her dissertation
Congratulations to Dr. Dan Liang, the first student to defend her Ph.D. dissertation in the lab. We’re so very proud of all you accomplished in graduate school!
Preprint on cell type specific eQTLs
Nil Aygün, a graduate student in the lab, has led a project to identify how genetic variation affects gene expression during human cortical neurogenesis. Importantly, this was completed in a cell-type specific approach, where we studied two cell types involved in neurogenesis - neural progenitors and their neuronal progeny. Most eQTLs were cell-type specific and not detected in previous bulk tissue eQTLs. This implies we may be missing a lot of gene regulation with bulk tissue QTLs. Also, many of these loci co-localize with neuropsychiatric disorder GWAS, allowing inference of genes dysregulated in these disorders. Check out the pre-print here.
Paper published in Human Brain Mapping on genetic architecture of brain traits as compared to neuropsychiatric disorders
Nana Matoba, a postdoc in the lab, led a study to evaluate whether brain traits satisfy one aspect of being an endophenotype for neuropsychiatric disorders. That is, are brain structure traits “closer to the underlying biology” than heterogeneous and behaviorally defined neuropsychiatric disorders? We evaluated this by comparing polygenicity, the total number of variants that are associated with a disorder, and discoverability, the distribution of effect sizes for a trait. An endophenotype is hypothesized to have reduced polygenicity and increased discoverability compared to a disorder. For some brain structure traits, like cortical surface area, relative to aggregated estimates across multiple neuropsychiatric disorders, we provide evidence to support brain structure traits as satisfying this aspect of an endophenotype. You can read more about this here.
NuMorph: analyze your tissue cleared images
Oleh Krupa led a project to develop a tool used for analyzing tissue cleared images. He imaged WT and Top1 cKO mice, getting beautiful cellular resolution images of the entire brain. Using the tool he developed, he was able to accurately quantify all nuclei in the cortex, accurately count individual cell types, and identify differences in cell type proportions and density between the genotype groups. This is a tool that we hope will be widely useful to the research community for studying genetically mediated differences in brain structure!
Preprint on identifying causal genes associated with traits
Both GWAS and eQTLs often have allelic heterogeneity - multiple independent loci close to each other influence the same trait/gene. You can use this information as a natural experiment to infer causality of a gene in trait levels. Do slight wiggles in levels of a gene expression lead to slight wiggles in the level of a trait? If so, that’s evidence for causal function of the gene in the levels of the trait. Led by Mike Love, we developed a tool called MRLocus that integrates eQTL and GWAS to identify causal genes mediating trait differences. Read more about it here: https://www.biorxiv.org/content/10.1101/2020.08.14.250720v2.full.
Jordan Valone starts grad school at UNC
Jordan Valone, a research technician who has been working with us for about a year, started graduate school today here at UNC in the BBSP program. Good luck Jordan and thanks for all of your work!
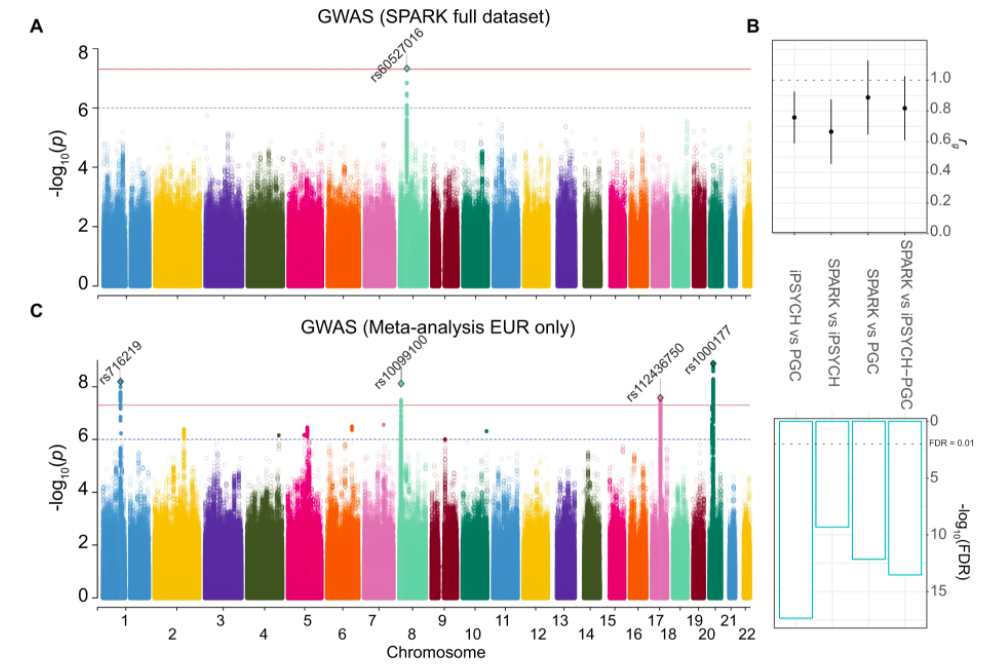
GWAS of Autism Spectrum Disorder in SPARK published in Translational Psychiatry
We completed a GWAS in the SPARK dataset , identifying a new genome-wide significant locus and performing some validation studies to identify causal variants within an associated locus. The work was published in Translational Psychiatry: https://www.nature.com/articles/s41398-020-00953-9.